PaccMannRL on SARS-CoV-2: Designing antiviral candidates with conditional generative models
Published in arXiv, 2020
Recommended citation: Born, Jannis et al., (2020). "PaccMannRL on SARS-CoV-2: Designing antiviral candidates with conditional generative models." arXiv2005(13285) https://arxiv.org/abs/2005.13285
A drug discovery framework for antiviral small molecules against SARS-CoV-2:
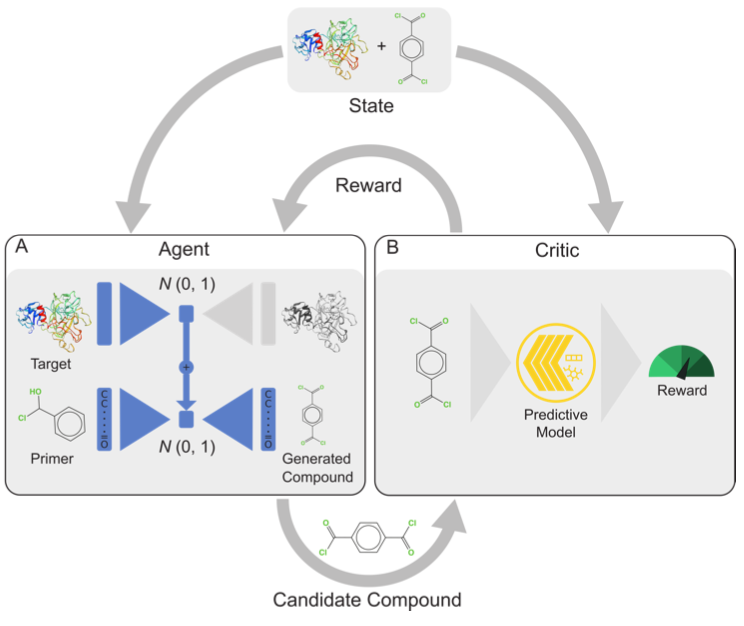 The conditional compound generator, called agent (see A), can produce novel structures specifically designed to target a protein of interest. The generative process starts with the encoding of the primary structure of the target protein into a latent space of protein sequences. The representation is fed into a molecular decoder of a separately pretrained molecule VAE to produce a candidate compound.
The conditional compound generator, called agent (see A), can produce novel structures specifically designed to target a protein of interest. The generative process starts with the encoding of the primary structure of the target protein into a latent space of protein sequences. The representation is fed into a molecular decoder of a separately pretrained molecule VAE to produce a candidate compound.
Next, the proposed compound is evaluated by a critic (see B) composed by: a multimodal deep learning model that predicts protein–drug binding affinity using protein and compound sequences as input, and a QSAR-based score to punish toxicity. By means of the reward given by the critic, a closed-loop system is created and is trained with deep reinforcement learning to maximize a multi-objective reward.
